Synapse Resolution Whole-Brain Atlases
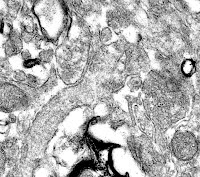
It is well-known that the highest resolution whole brain atlases are currently at BrainMaps.org, which has been compared to a Google Maps for the brain. However, these atlases are 0.46 microns per pixel, and are not sufficient to discern individual synapses, which require nanometer resolution. So in this post, I will consider the problems associated with constructing a synapse resolution (nanometer resolution) whole-brain atlas.
There seem to be two fundamental hurdles to constructing a synapse resolution whole-brain atlas: 1) image acquisition, and 2) digital technologies for working with the images and serving them over a network.
The first hurdle encompasses the time bottleneck and section preparation. If each section is 50 nm thick, then for a 10 mm mouse brain, 200,000 sections are needed, thus requiring some type of automation for section preparation. If we consider the time to scan a single 10mmx10mm section at 10nm pixel size at 1MHz, it comes out to 12 days (additional overheads would come from stage movements). Even with 200,000 TEMs (transmission electron microscopes) in parallel, one for each section, it will take 12 days for the complete scan. An alternative is offered by way of virtual microscopy solutions offered for light microscopy. One way would be to scan over the section, acquiring one column at a time instead of a patchwork of small images for montaging. Another alternative would be to construct a TEM with parallel scanning capabilities (having parallel magnetic lenses and electron beams), so that the entire section could be scanned at once, instead of scanning each little image patch in serial. This solution requires constructing a special type of TEM which implements certain features found in current day virtual microscopy systems for LM (light microscopy), and thus requires a team of hardware and software specialists to specially design, in addition to some physicists who are intimately acquianted with the physics behind TEM.
The second hurdle involves digital technologies, and the observation that even if a whole mouse brain was able to be acquired through TEM, that digital technologies currently would not be able to deal with that much data (200 PB, uncompressed). A single section is 1 x 10^12 pixels, which comes out to 1 TB (uncompressed, 8 bit grayscale), which is still not feasible using today's digital technologies.
In conclusion, for purposes of obtaining information about whole-brain connectivity, a nanometer-resolution whole-brain scan is required, and current-day tracer experiments are suboptimal and will always leave room for ambiguities that can only be resolved by completely mapping every synapse and axon in the brain. However, constructing a synapse resolution (or nanometer resolution) whole-brain atlas for even a mouse brain is so formidable as to be seemingly beyond today's technological capabilities. Maybe in 10-20 years.
Labels: brain atlas, digital technologies, electron microscopy, light microscopy, synapses, virtual microscopy
